NGS analysis for phage display and other display technologies
Phage display is a high throughput screening technology to identify protein ligands (e.g. a phage display antibody) or to study natural protein–protein or protein–DNA interactions. The phage display technology uses bacteriophages, which link the genetic information to various combinatorial libraries expressed on their surface. These libraries may be composed of peptides, antibodies, Fab fragments or other scaffolds. Further methods besides phage display are mRNA display, yeast display or bacterial display.
All these methods have in common that iterative cycles of selection and amplification are used to finally isolate the ligand of interest from the respective combinatorial library.The phage display technology for the identification of protein ligands:
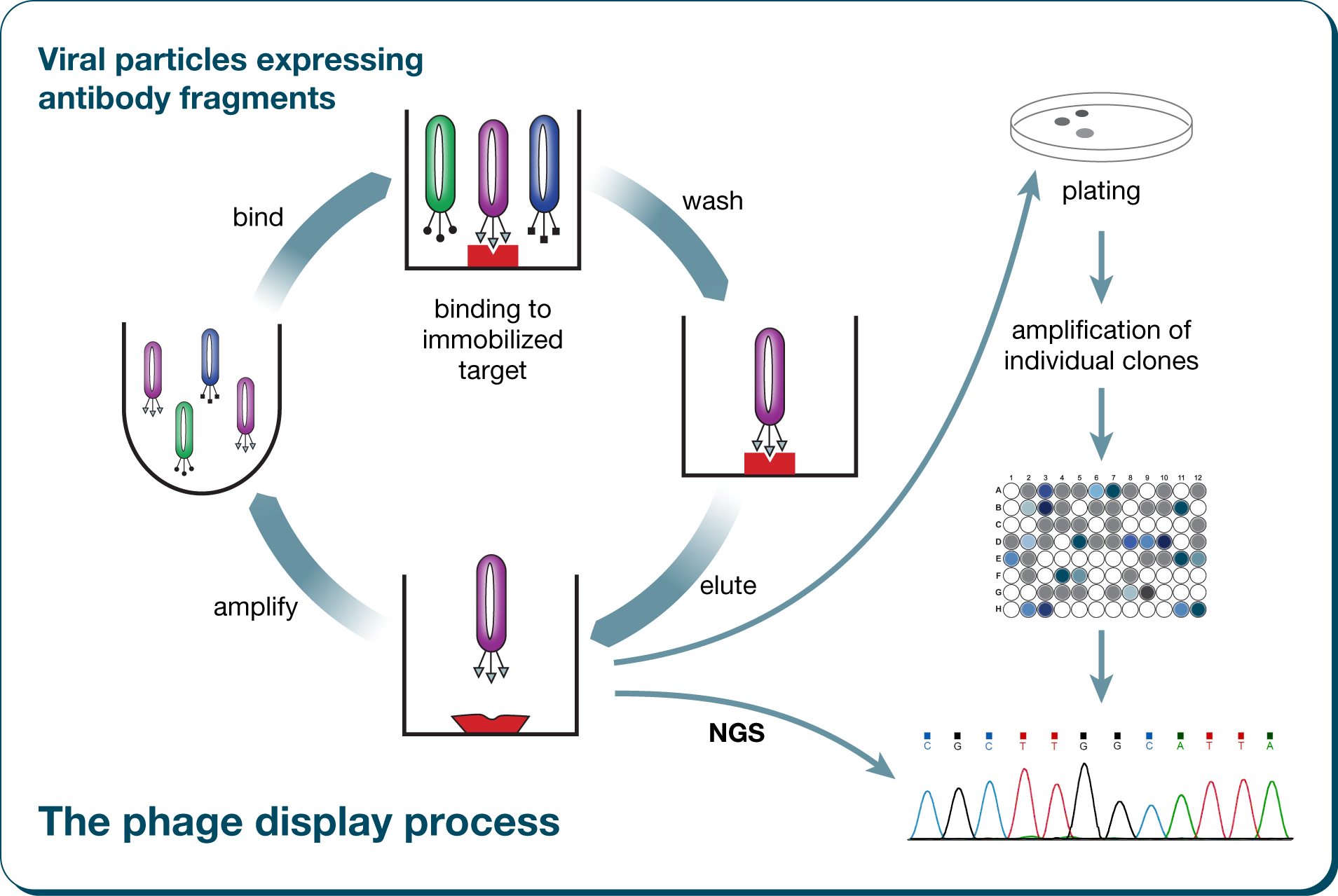
The technology of next-generation sequencing (NGS) is able to provide millions of sequences of libraries or defined selection cycles within a very short time. This allows for the first time an unprecedented deep insight into the evolutionary processes within an individual biopanning experiment. Our software AptaAnalyzer™-Peptide supports the complete workflow of next-generation sequencing data analysis in a very intuitive way. It is one tool for parsing of raw data, archiving of datasets, analysis of experiments, and generation of results in form of table views and publication level graphics.
NGS data analysis for phage display experiments:
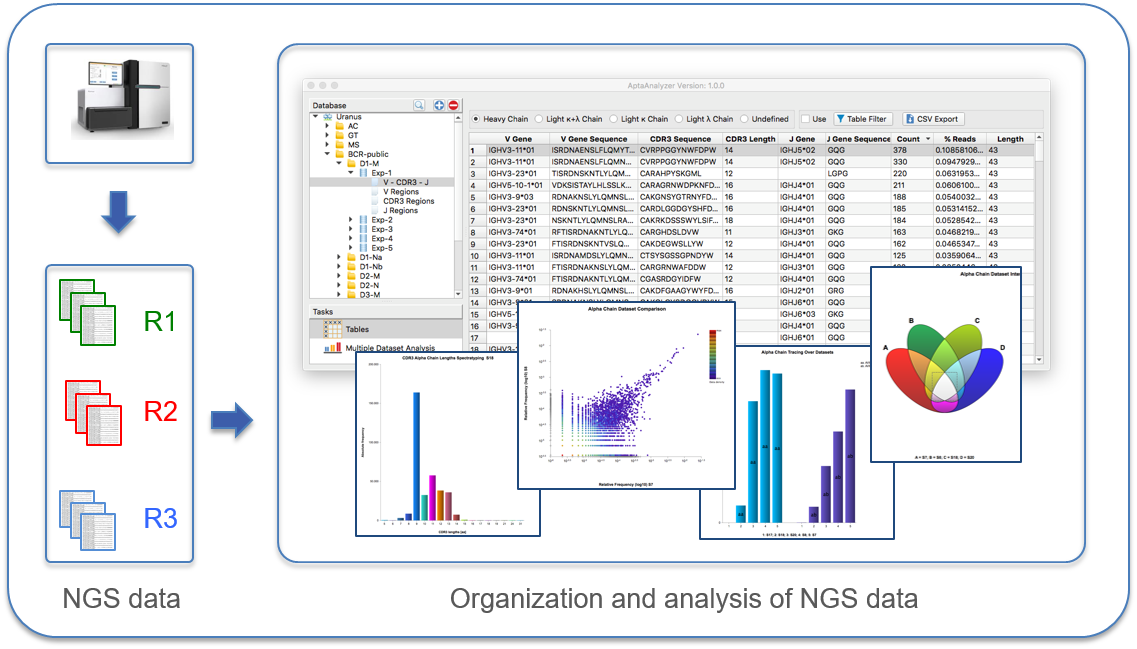
Therefore, NGS is part of innovative phage or other display methods, which can accelerate the following steps:
- Quality control of combinatorial phage display libraries
- More effective identification of ligands, e.g. phage display antibodies
- Optimization of phage display lead candidates
Quality control of phage display libraries
The outcome of biopanning experiments is strongly dependent the quality of the initial phage display library , which is used as the source to extract the best binders. The more the distribution of amino acids is in accordance with the specifications, the better is the library’s diversity and the chance to select better binders. Next-generation sequencing of the initial phage display libraries is a very helpful method to receive solid statistical data on the distribution of amino acids in defined positions or frequencies of motives in a starting library. The sequencing data can be examined on the level of nucleotides as well as amino acids.
Identification of phage display ligands
Analyzing selection or panning rounds in a phage display experiment by next-generation sequencing yields millions of sequence candidates. AptaIT’s bioinformatics approach provides the necessary tools get comprehensible results within a short time period and can be applied to virtually any other display technology . NGS helps to solve several bottlenecks of the phage display technology:
- Selecting promising candidates within the thousands of sequences after just one round of panning
- Subtracting false positives and contaminations
- Successfully applying difficult selections, e.g. negative selections, competitive elution, or cells as targets
- Tracking ligands over comparative selection experiments
Optimization of phage display lead candidates
Following the identification of ligand families with the expected properties in a phage display experiment, the already available NGS sequence information contained in the entire family can be used to define conserved and optimized binding motives, which is invaluable information with respect to phage display derived antibody or peptide lead optimization. The almost unlimited data of functional binding motives and their variations additionally allow comprehensive patenting strategies.
Download the phage display application note for a more detailed overview. AptaIT is providing innovative solutions to help researchers to exploit the full potential of next-generation sequencing data analysis: Our easy-to-use software AptaAnalyzer™-Peptide and custom NGS data analysis services.
